Written by Andreas Maier.
In the age of deep learning, data became an important resource to build powerful smart systems. In several fields, we already see that the amount of data that is required to build competitive systems is so large that it is virtually impossible for new players to enter the market. For example, state-of-the-art large vocabulary speech recognition systems that are available from major players such as Google or Nuance are trained with up to 1 million hours of speech. With such large amounts of data, we are now able to train speech-to-text systems with accuracies of up to 99.7%. This is close to or even exceeds the human performance, given that the system does not need breaks, sleep, or ever gets tried.

Besides the collection, the data also needs to be annotated. For the speech example, one hour of speech data requires approximately 10 hours of manual labor to write down every word and non-verbal event such as coughs or laughs. Hence, even if we had access to 1 million hours of speech, the transcription alone neglecting the actual software development cost – given a $5 hourly rate – would equal a $50 million investment. Hence, most companies prefer to license a state-of-the-art speech recognition system from one of the current software suppliers.
For the case of medical data, things are even more complicated. Patient health data is – for good reasons – well protected by patient data laws. Unfortunately, the standards differ considerably from country to country which makes the issue even more complicated. Lately, several big hospitals, companies, and health authorities made data publicly available in an anonymized way in order to drive deep learning research ahead. Still those datasets only reach counts from several 10s to several thousand and the associated annotations generally show significant variation, as annotations are typically only done once per dataset.
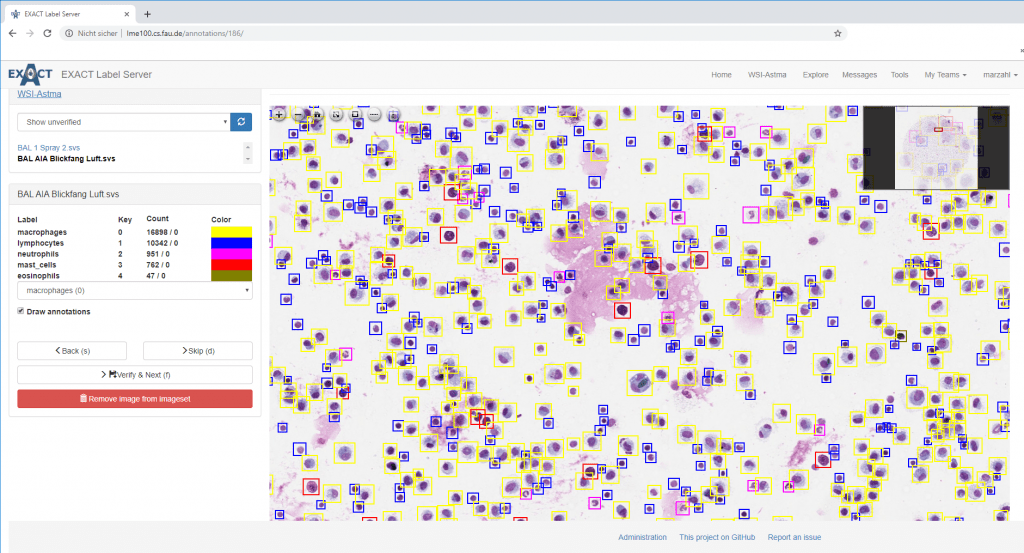
In particular in medical image analysis, these public datasets are extremely useful to drive current research ahead. As we have seen in speech processing such smaller datasets (for speech approx 600 hours) are suited to develop good software to approach the task. In speech processing, these systems were able to recognize 90-95% of the spoken words. The game-changer that made 99.7% possible, however, was the 1 million hours of speech data.
This observation leads to the requirement that we will need at some point millions of well-annotated training images to build state-of-the-art medical analysis systems. There are very few methods to actually achieve this goal: Significant investment by big industry players, organization via government authorities, or non-government organizations. One exception may be digital pathology where public data can be generated from animal specimens.
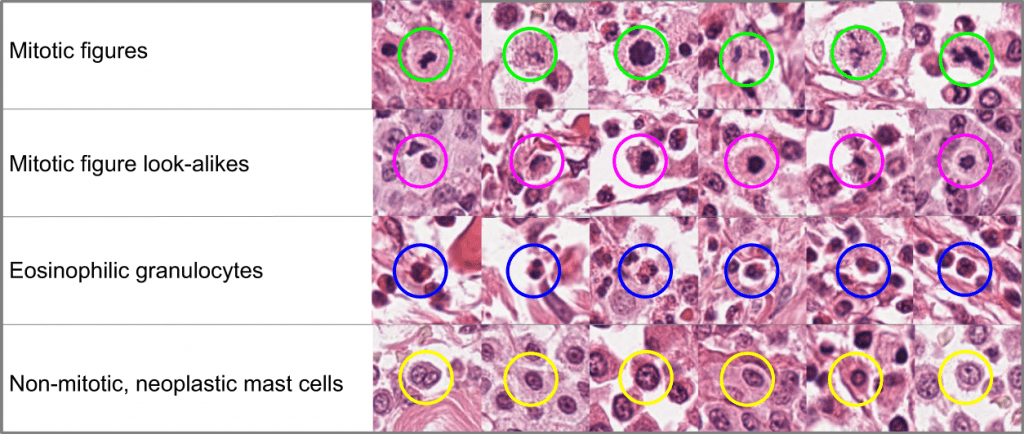
While speech and other machine learning training data are already predominantly controlled by industries, one may ask whether we want the same happen to our medical records. There are good reasons why these data are well protected and are e.g. not sold to insurance companies without our knowledge. So each of us should ask her- or himself whether this is a reasonable solution or not.
Some countries are actually already starting to process medical data in government-controlled databases that allow access to researchers and industrial developments. Denmark is an example that is already following this path. It will be interesting to see future developments happening in Denmark and other countries.

Only this year, a small non-profit organization was founded in Germany called „Medical Data Donors e.V.“. They follow the third path and ask patients to donate image data for research and development. Following the new European data protection guidelines, they impose high ethical standards. Even within this strong framework of supervision, they are able to collect and share data worldwide. While this effort is only starting and the organization is only small, it will be interesting to see how far they can get. This is in particular interesting, as they attempt at solving the annotation problem by gamification. A storyboard for the game is already available. Hence, they would not just collect data, but also generate high-quality annotations.
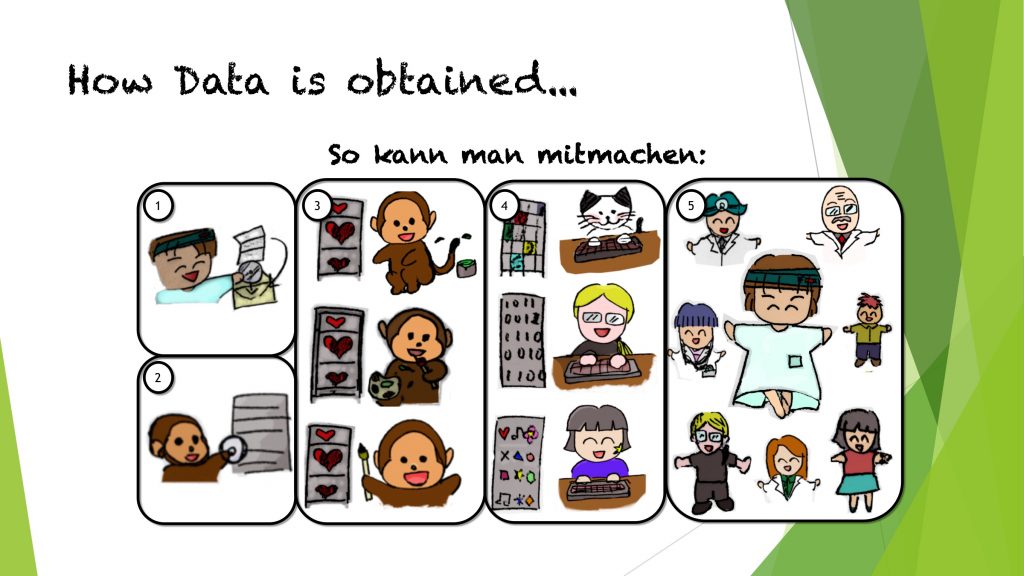
Only recently, Medical Data Donors published further ideas how actual games for data annotation could look like. They adopt common games such as racing for the aim of organ outlining or candy crush to assign labels to images. In particular, Odin’s Eye seems to be an exciting approach for crowd annotation of ophthalmic image data.
In summary, we see that the medical data problem is far from being solved. We identified three different feasible solutions to attack the problem: industrial investment, state control, or non-government organisations. While all of them are possible, we have to ask ourselves which ones we prefer. In any case, the issue is urgent and needs to be solved in order to push deep learning research in medicine ahead.
If you liked this essay, you can have a look at my YouTube Channel. This article originally appeared first on MarkTechPost.com and is released under the Creative Commons 4.0 Attribution License.

