Thesis Description
The advent of deep learning in recent years has led to a multitude of new practical applications of machine learning in many fields, including the medical domain [1]. A main limiting factor in
the implementation of deep learning algorithms for healthcare applications is the availability of representative training datasets of sufficient size [2].
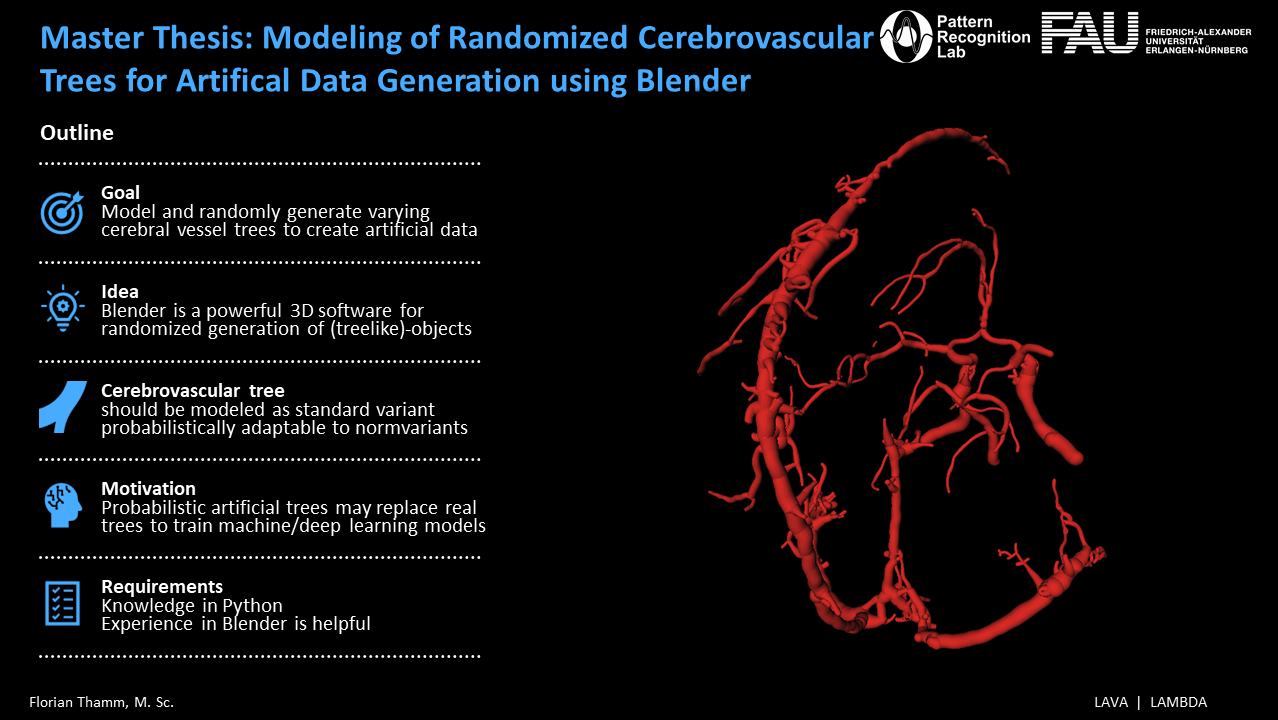
Solving the data scarcity problem may prove particularly beneficial in the case of stroke. Globally, stroke is the leading cause of serious adult disability and the second-leading cause of death [3]. The main structure to distribute blood flow to the brain is the Circle of Willis (CoW) [3, 4]. Several anatomical variations of the CoW can be observed in the population [3, 4]. These normvariants differ in the frequency they appear in, leading to only 40% of the population possessing a wellformed complete CoW [4].
The multitude of CoW normvariants further exacerbates the need for more cerebrovascular training data for tasks like vessel labeling in stroke cases [5]. It may be possible to alleviate this problem by generating artificial data. The open-source 3D graphics software Blender appears suitable for this.
The aim of this thesis is to model a CoW graph which can be randomly and realistically deformed while being able to probabilistically incorporate common normvariants. The thesis shall comprise the following points:
- Literature research regarding the distribution of CoW normvariants in the population
- Model a standard variant of the cerebrovascular tree in Blender
- Create artificial trees by randomly sampling its parameter values
- Probabilistically adapt model to normvariants
References
[1] Neha Sharma, Reecha Sharma, and Neeru Jindal. Machine learning and deep learning applications-a vision. Global Transitions Proceedings, 2(1):24–28, 2021. 1st International Conference on Advances in Information, Computing and Trends in Data Engineering (AICDE – 2020).
[2] Martin J. Willemink, Wojciech A. Koszek, Cailin Hardell, Jie Wu, Dominik Fleischmann, Hugh Harvey, Les R. Folio, Ronald M. Summers, Daniel L. Rubin, and Matthew P. Lungren. Preparing medical imaging data for machine learning. Radiology, 295(1):4–15, 2020.
[3] Mohammed Oumer and Mekuriaw Alemayehu. Association between circle of willis and ischemic stroke: a systematic review and meta-analysis. BMC Neuroscience, 22(10), 2021.
[4] Debanjan Mukherjee, Neel D. Jani, Jared Narvid, and Shawn C. Shadden. The role of circle of willis anatomy variations in cardio-embolic stroke – a patient-specific simulation based study.
bioRxiv, 2018.
[5] Florian Thamm, Markus J¨urgens, Hendrik Ditt, and Andreas Maier. VirtualDSA++: Automated Segmentation, Vessel Labeling, Occlusion Detection and Graph Search on CTAngiography Data. In Barbora Kozl´ıkov´a, Michael Krone, Noeska Smit, Kay Nieselt, and Renata Georgia Raidou, editors, Eurographics Workshop on Visual Computing for Biology and Medicine. The Eurographics Association, 2020.