Description
The task of segmentation of organs or lesions in medical images such as magnetic resonance or computed tomography plays an important role for clinicians. Based on the annotations, volumes and measures can be made that characterize the patient’s disease state and track it over time. However, manual, or semi-automatic segmentation takes a substantial amount of time, requires experienced annotators, and in some cases has large inter-reader variance. Instead of manually annotating large amounts of data by hand, machine learning-based methods allow to propose segmentations without further attention required. Deep learning-based neural networks have achieved outstanding performance for cell segmentation, organ segmentation, and cardiac segmentation. However, prior information about the organ of interest is typically harder to incorporate into deep learning models. Previous works utilized active shape models (ASM) or relied on the principal component analysis to enforce constraints extracted from prior information [4,5].
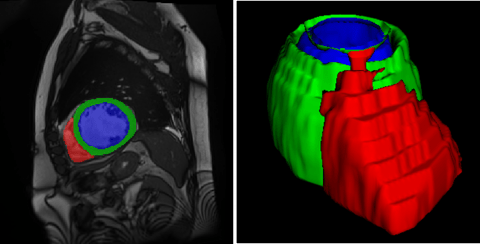
In this thesis, spatial transformer networks (STN) [1] will be utilized in combination with reference shapes to obtain predictions heavily utilizing prior information. The dataset proposed for this task consists of 100 three-dimensional volumes of the heart [2] (https://acdc.creatis.insa-lyon.fr/).
 [2]
[2]
Steps
This thesis can be divided into the following steps:
- Training of a segmentation network (e.g., nn U-Net [3]).
- Application of a rigid 2D STN to the prediction of the segmentation network + reference shape.
- Extension to deformable 2D STN.
- Extension of STN to 3D input and application to the segmentation prediction.
Figure 1: Example case of the ACDC dataset with overlaying segmentation masks (left) and 3D rendering of the segmentation mask (right) [2].
Requirements
- Good understanding of machine learning/deep learning concepts
- Strong knowledge of Python and ideally Pytorch is required
References
[1] Jaderberg, Max, et al. “Spatial transformer networks.” arXiv preprint arXiv:1506.02025 (2015).
[2] O. Bernard, A. Lalande, C. Zotti, F. Cervenansky, et al. “Deep Learning Techniques for Automatic MRI Cardiac Multi-structures Segmentation and Diagnosis: Is the Problem Solved ?” in IEEE Transactions on Medical Imaging, vol. 37, no. 11, pp. 2514-2525, Nov. 2018
[3] Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J., & Maier-Hein, K. H. (2020). nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods, 1-9.
[4] Milletari F., Rothberg A., Jia J., Sofka M. (2017) Integrating Statistical Prior Knowledge into Convolutional Neural Networks. In: Descoteaux M., Maier-Hein L., Franz A., Jannin P., Collins D., Duchesne S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science, vol 10433. Springer, Cham.
[5] Ahmadi, S.-A., Baust, M., Karamalis, A., Plate, A., Boetzel, K., Klein, T., Navab, N.: Midbrain segmentation in transcranial 3D ultrasound for parkinson diagnosis. In: Fichtinger, G., Martel, A., Peters, T. (eds.) MICCAI 2011. LNCS, vol. 6893, pp. 362–369. Springer, Heidelberg (2011)