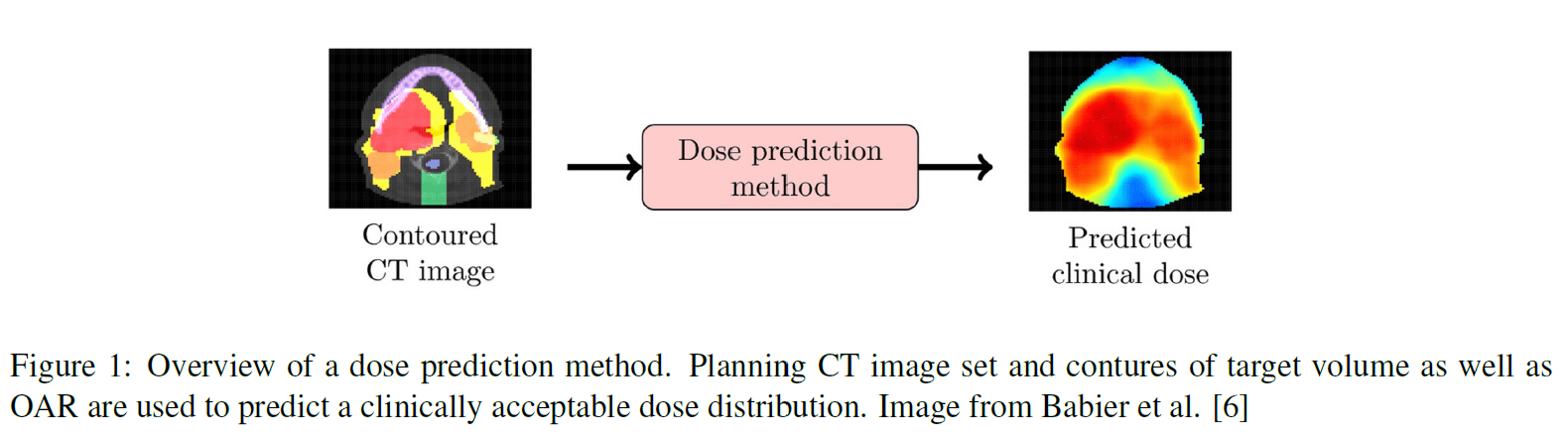
Radiation therapy is one of the most important local cancer treatment modalities, enabling the non-invasive delivery of a spatially varying dose distribution within a patient’s body with high precision. Radiotherapy treatment planning aims to identify the optimal treatment plan that maximally spares surrounding normal tissue structures while delivering a high dose to the target volume. Deep Learning dose prediction is a novel technique that can automate the treatment planning process, improve standardization and reduce the treatment planning time to enable novel and improved therapies like same-day treatment and adaptive radiotherapy [1]. Deep learning dose prediction uses deep learning models like 3D U-Nets [2] to predict spatial dose distributions based on planning CT datasets, target volume, and organs at risk (OAR) segmentations [1, 3] (Figure 1). Subsequently, dose mimicking algorithms are used to translate these 3D dose predictions into deliverable treatment plans [1, 3–5]. An additional resource for dose prediction research is provided by the Open Knowledge-Based Planning Challenge (OpenKBP) [6], which offers a publicly available dataset and a standardized evaluation framework. The aim of this master thesis project is to improve an existing 3D
U-Net dose prediction model and evaluate its performance using a cohort of more than 450 Head & Neck cancer datasets from the Department of Radiation Oncology, University Hospital Erlangen.
The thesis will include the following points:
• Literature review on deep learning techniques for radiotherapy dose prediction and automated treatment planning.
• Data preprocessing for the private Head & Neck treatment plan dataset including conversion of DICOM (CT, RT DOSE, RT STRUCT) files into CSV files and label maps. Automatic cleaning and splitting of the datasets into subgroups according to plan parameters using custom Python scripts (Pydicom library).
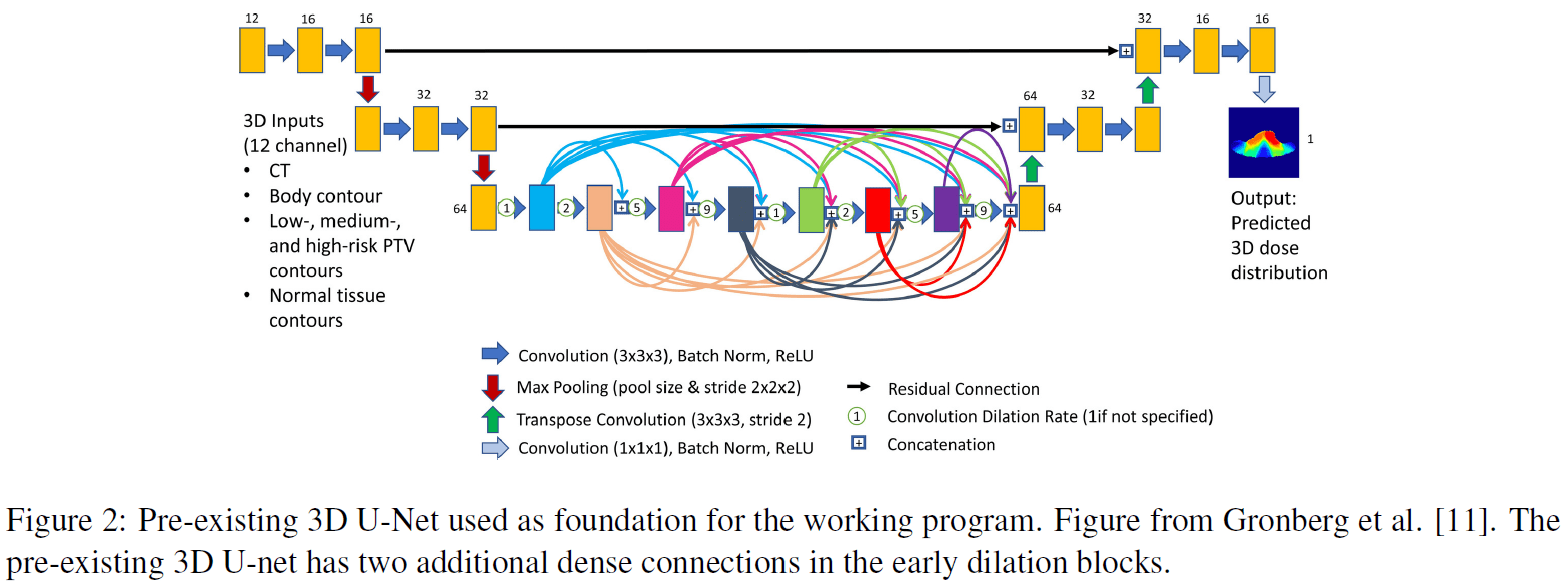
• Training, Inference and Testing of a pre-existing 3D U-net (Figure 2) dose prediction model (Keras, PyTorch) on the cohort of 450 Head & Neck cancer datasets.
• Reframe of the pre-existing dose prediction model to enable multi-GPU training with PyTorch. Enable increased resolution of the predicted voxel grid using multi-GPU training, splitting into subvolumes and/or further strategies to decrease memory consumption (e.g., Automatic Mixed Precision). Optimization of training time and GPU usage by shifting pre-processing steps to before the training phase.
• Extension of the pre-existing dose prediction model by implementing an on-line augmentation pipeline tailored to the task of dose prediction. Hyperparameter optimization on the validation dataset.
• Optional: Exploration of additional deep learning architectures for radiotherapy dose prediction including novel techniques like diffusion models [7, 8] and transformer architectures [9, 10].
• Optional: Adaption of the developed dose prediction pipeline to lung tumors with a data set of 130 patient plans.
• Detailed evaluation of the deep learning dose prediction performance on the test dataset comparing automated treatment plans to manually created treatment plans. Comparison of the different deep learning approaches in regard to accuracy and inference time. Five training and inference repeats to enable statistical analysis.
If you are interested in the project, please send your request to: johann.brand@uk-erlangen.de
Having prior experience with building neural networks in Python, especially using frameworks such as PyTorch or TensorFlow, will greatly help to develop the project.
